About the author: Trevor Ghylin is founder and chief technology officer of Microbe Detectives. Ghylin can be reached at [email protected].
Industrial wastewater facilities tend to treat wastes with highly variable flows and concentrations. As a result, these facilities tend to be difficult to operate and may have issues with settling, toxicity and ammonia removal. Operators struggle to understand operational issues due to a lack of information about the bacteria responsible for these processes. Troubleshooting issues may consist of microscopy as well as checking pH, alkalinity, MLSS and a lot of head scratching. The key limiting factor is our inability to see into the microbial world of these systems.
DNA Discovery
Traditionally, biological monitoring and troubleshooting of activated sludge systems has been performed using a microscope, not unlike microscopes that have been around for more than 100 years. The microscope is able to give us some very quick information (i.e. filaments, pin floc, protozoans, etc.), but does not provide precise identification or quantification of beneficial or problem bacteria.
A very large body of knowledge has been built based on microscopy to troubleshoot settling issues; however, this method requires a trained observer and is limited only to organisms that have unique shapes and staining features. The vast majority of microbes in activated sludge do not have unique shapes or staining features, including some of the most critical bacteria like nitrifiers (ammonia removal) and phosphate accumulating organisms. As a result, microscopy provides essentially no information for troubleshooting issues with ammonia or phosphorus removal.
Recent advances in DNA sequencing allow us to sequence DNA from samples of biomass in wastewater treatment plants and identify and quantify nearly all the microbes in the system. This allows us to provide precise identification and quantification of all the microbes important to the system. Now that we finally can “look inside the black box,” we can start to solve problems and optimize the system.
More Than Data
Wastewater treatment relies on microbiological processes, such as activated sludge, with little knowledge of the microbial community present. Instead, these systems are typically operated based on empirical monitoring data, such as dissolved oxygen and effluent BOD.
This simple operational strategy generally works for plants most of the time. However, problems can arise when plants are stressed due to high loading, low temperatures, toxic influent, and growth of filaments and foam producing organisms. These problems are especially prevalent in industrial treatment facilities where influent can be variable and difficult to treat.
Beyond the Microscope
The traditional method of troubleshooting activated sludge consists of analyzing a droplet of sludge under a microscope. This technique is easy and cheap and can provide some information very quickly. However, it has significant limitations because it cannot identify most microbes that are important to treatment [e.g., nitrifiers, phosphorus removers (PAOs)].
Microscope users attempt to identify problematic filaments and foaming organisms; however, this task may be impossible as the problem organisms can be hidden inside flocs, and many different species of filaments and foaming organisms have identical morphologies. In addition, morphologies can change under varied nutrient conditions. Due to the difficulties in microscopic characterization, many studies have focused on creating fluorescent molecular probes that bind to the DNA of the target organism and fluoresce to provide visual identification under a microscope. In this way, organisms with identical morphology can be distinguished based on their DNA.
FISH
Many probes have been developed to target common filaments, nitrifiers and PAOs. However, there are so many organisms that create filaments and foaming that it is difficult to identify problem organisms in a sample with this method. A 2002 study, Thomsen et al, 2002, showed that Fluorescent In-situ Hybridization (FISH) probes were only able to identify 15% of organisms in a foam sample, and these organisms were only identified at a very high taxonomic level (phylum). The FISH method also can suffer from inefficient permeabilization of cells, inefficient binding to DNA and lack of specificity. In addition, it can be somewhat complex, sometimes requiring multiple helper probes or special methods to increase the fluorescent signal-like catalyzed active reported deposition (CARD)-FISH. The FISH method is not something that a typical wastewater treatment lab can perform routinely as it requires some biotechnology expertise and an epifluorescence microscope. Additionally, there are no companies currently offering this as a commercial service. As a result, this method is only appropriate for an academic laboratory with experience in DNA methods.
DNA Sequencing
A more recent advancement is next generation DNA sequencing. With DNA sequencing, we are able to characterize entire microbial communities based on the DNA of individual organisms with much less complexity and potential problems than FISH. This method is able to provide quantification of nitrifiers, PAOs, filaments, foamers and other organisms in a sample. D
NA technologies have been exponentially decreasing in cost since the first human genome was published in 2001 at a cost of $3 billion (Figure 1, page 11). The cost of sequencing a human genome has dramatically fallen. The same technology that is used to sequence human genomes can be used to analyze activated sludge for a similar price as sending a sample for microscopic analysis of filaments.
The cost of these technologies are at the point where it has become economically feasible to identify and quantify nearly every microbe in a sample of wastewater using DNA.
Employing DNA
A wastewater treatment plant at a large food manufacturer experienced frequent upsets that caused the sludge volume index (SVI) to soar higher than 300, resulting in poor sludge compaction in the clarifier and high effluent total suspended solids (TSS).
Although permit violation was not an issue, the excessive discharge load resulted in massive sewer charges surpassing $50,000 per month. Four samples were collected from the treatment facility to diagnose the issue. A sample was collected from the moving bed bioreactor just upstream of the activated sludge system. Samples were also collected from the activated sludge basin, the clarifier effluent and the clarifier return activated sludge (RAS). Analysis was performed to compare the bacteria in the clarifier effluent to those found in the RAS.
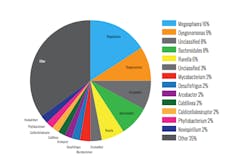
In this way, Microbe Detectives could determine which bacteria were settling well and which were settling poorly (Tables 1 and 2, page 12). Suprisingly, two well known filaments—haliscomenobacter and sphaerotilus—were some of the best settling bacteria. The poor settling bacteria included aureispira, of which little is known, and zoogloea, a well-known bulking bacteria usually present due to nutrient limitations.
The differences between the clarifier and the RAS were not as great as expected, indicating that the mixed liquor suspended solids (MLSS) is relatively homogenous and all of it settles poorly. Attention turned to the most abundant bacteria in the MLSS to find the culprits for the high SVI. Table 3 (page 12) shows that the most abundant bacteria include aureispira, as well as several known filaments—sphaerotilus, thiothrix, haliscomenobacter—and zoogloea.
Based on the DNA data and the presence of zoogloea, the plant will further investigate nutrient deficiency—particularly nitrogen or phosphorus—that may contribute to bulking, causing high SVI, high effluent TSS and high sewer bills.
Power to the Plant
DNA sequencing is a powerful method to troubleshoot problems in wastewater treatment plants. This tool is especially powerful when operators have baseline samples analyzed during healthy operation to provide a comparison with poor operation. The spreadsheet format of this data makes it easy to analyze and compare across historical data. DNA data can be obtained by simply shipping off a small sample of activated sludge. This data may provide the final piece of the puzzle to solve longstanding, recurring issues with foaming, filaments, nitrifiers or PAOs.